Exploring the relationship between two variables
First lets bring in the data from the previous lesson
library(tidyverse)
library(broom)fish_data <- read_csv("https://raw.githubusercontent.com/chrischizinski/MWFWC_FishR/master/CourseMaterial/data/wrkshp_data.csv")## Parsed with column specification:
## cols(
## .default = col_character(),
## WaterbodyCode = col_integer(),
## Area = col_integer(),
## MethodCode = col_integer(),
## surveydate = col_datetime(format = ""),
## Station = col_integer(),
## Effort = col_integer(),
## SpeciesCode = col_integer(),
## LengthGroup = col_integer(),
## FishCount = col_integer(),
## FishLength = col_integer(),
## FishWeight = col_integer(),
## Age = col_integer(),
## Edge = col_integer(),
## Annulus1 = col_integer(),
## Annulus2 = col_integer(),
## Annulus3 = col_integer(),
## Annulus4 = col_integer(),
## Annulus5 = col_integer(),
## Annulus6 = col_integer(),
## Annulus7 = col_integer()
## # ... with 2 more columns
## )## See spec(...) for full column specifications.fish_data %>%
select(WaterbodyCode:Age) %>%
mutate(Age = as.numeric(Age)) %>%
filter(!is.na(Age),
WaterbodyCode == 4999,
SpeciesCode %in% c(780)) -> FishAge Now lets plot the basic relationship between age and length of this species.
ggplot(data = FishAge) +
geom_point(aes(x = Age, y = FishLength), size = 3, alpha = 0.35, colour = "red") +
theme_bw()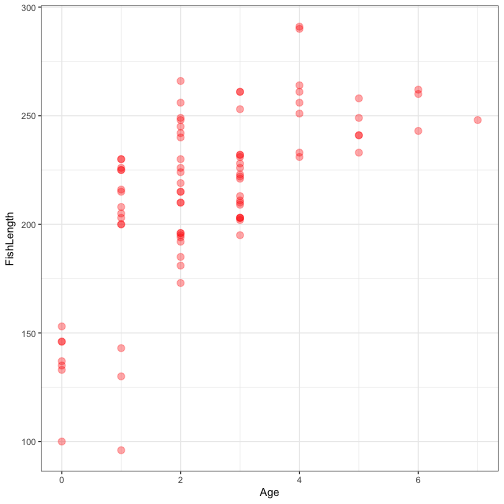
How does fish age relate to fish length?
- Linear
- Polynomial
- Logarithmic
- Non-linear
# linear response
lm_mod <- lm(FishLength ~ Age, data = FishAge)
summary(lm_mod)##
## Call:
## lm(formula = FishLength ~ Age, data = FishAge)
##
## Residuals:
## Min 1Q Median 3Q Max
## -94.265 -19.517 0.703 21.953 58.725
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 173.254 6.232 27.799 < 2e-16 ***
## Age 17.011 2.138 7.957 9.8e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 29.81 on 80 degrees of freedom
## Multiple R-squared: 0.4418, Adjusted R-squared: 0.4348
## F-statistic: 63.32 on 1 and 80 DF, p-value: 9.797e-12# polynomial response
poly_mod <- lm(FishLength ~ poly(Age,3), data = FishAge)
summary(poly_mod)##
## Call:
## lm(formula = FishLength ~ poly(Age, 3), data = FishAge)
##
## Residuals:
## Min 1Q Median 3Q Max
## -92.393 -18.407 -1.423 19.566 49.441
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 215.366 3.011 71.527 < 2e-16 ***
## poly(Age, 3)1 237.206 27.266 8.700 4.13e-13 ***
## poly(Age, 3)2 -107.773 27.266 -3.953 0.000169 ***
## poly(Age, 3)3 38.539 27.266 1.413 0.161501
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 27.27 on 78 degrees of freedom
## Multiple R-squared: 0.5447, Adjusted R-squared: 0.5272
## F-statistic: 31.1 on 3 and 78 DF, p-value: 2.506e-13# logarithmic
log_mod <- lm(FishLength ~ log(Age +1), data = FishAge)
summary(log_mod)##
## Call:
## lm(formula = FishLength ~ log(Age + 1), data = FishAge)
##
## Residuals:
## Min 1Q Median 3Q Max
## -93.378 -17.928 -2.378 18.426 52.822
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 148.691 7.462 19.925 < 2e-16 ***
## log(Age + 1) 58.699 6.023 9.745 3.04e-15 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 26.98 on 80 degrees of freedom
## Multiple R-squared: 0.5428, Adjusted R-squared: 0.5371
## F-statistic: 94.97 on 1 and 80 DF, p-value: 3.039e-15# non linear
nl_mod <- nls(FishLength ~ exp(a + Age*b), start = list(a = 0, b = 1), data = FishAge)
summary(nl_mod)##
## Formula: FishLength ~ exp(a + Age * b)
##
## Parameters:
## Estimate Std. Error t value Pr(>|t|)
## a 5.191797 0.031178 166.522 < 2e-16 ***
## b 0.070687 0.009528 7.419 1.1e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 30.64 on 80 degrees of freedom
##
## Number of iterations to convergence: 7
## Achieved convergence tolerance: 0.00000223Plot the curves to the data
newdata <- data.frame(Age = seq(0,8,by = 1))
lm_pred<- data.frame(model = "linear",augment(lm_mod, newdata = newdata))
poly_pred<- data.frame(model = "polynomial",augment(poly_mod, newdata = newdata))
log_pred<- data.frame(model = "log",augment(log_mod, newdata = newdata))
# log_pred$.fitted <- exp(log_pred$.fitted)
nl_pred<- data.frame(model = "nonlinear",augment(nl_mod, newdata = newdata))
nl_pred$.se.fit<-NA
all.pred<- rbind(lm_pred,poly_pred,log_pred,nl_pred)
ggplot(data = FishAge) +
geom_point(aes(x = Age, y = FishLength), size = 1, alpha = 0.35, colour = "red") +
geom_line(data=all.pred, aes(x = Age, y = .fitted, colour = model)) +
theme_bw()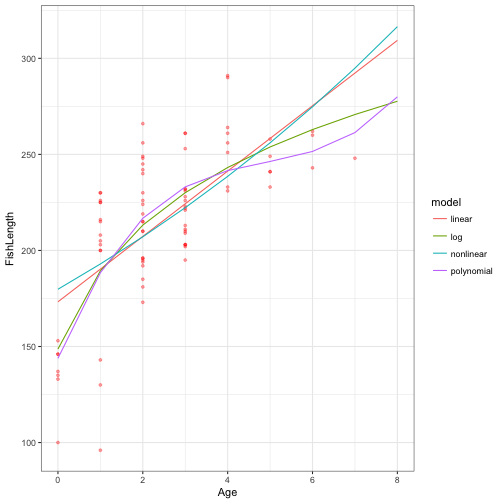
Lets look at the distribution of the residuals of each model to the actual data. What would you expect to see?
resid_lm<-data.frame(model = "linear",resid = augment(lm_mod)$.resid)
resid_log<-data.frame(model = "log",resid = augment(log_mod)$.resid)
resid_poly<-data.frame(model = "poly",resid = augment(poly_mod)$.resid)
resid_nl<-data.frame(model = "nonlinear",resid = augment(nl_mod)$.resid)
all_resid<-rbind(resid_lm, resid_log, resid_poly, resid_nl)
head(all_resid)## model resid
## 1 linear -44.3284415
## 2 linear 7.7139542
## 3 linear -12.2754468
## 4 linear 22.7033553
## 5 linear -13.3178426
## 6 linear -0.3072437glimpse(all_resid)## Observations: 328
## Variables: 2
## $ model <chr> "linear", "linear", "linear", "linear", "linear", "linea...
## $ resid <dbl> -44.3284415, 7.7139542, -12.2754468, 22.7033553, -13.317...ggplot(data = all_resid) +
geom_violin(aes(x = model, y =resid, fill = model)) +
theme_bw()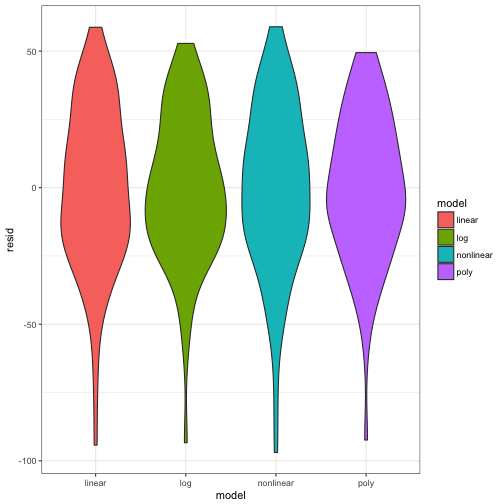
Experiments, Events, and Probability
-
In probability theory, we are concerned with the occurence of events that can be thought of as outcomes of experiments
-
The probability of event A occurring is \( Pr { A } \) = probability that the event occurs
-
The Frequentist interpretation of probability \( Pr { A } \) is the proportion of A outcomes as the total number of trials in an experiment goes to infinity.
Coin flipping example:
For example, it can be demonstrated that the proportion of heads from a series of fair coin flips will approach the constant 0.5 as the number of trials grows large, that is, \( Pr { Head } \) = 0.5
## Binomial distribution
?rbinom
set.seed(12345)
N_flips<-data.frame(N = c(1:5000))
N_flips$N_heads <- unlist(lapply(lapply(N_flips$N, rbinom, size = 1, prob = 0.50), sum))
N_flips$prop <- N_flips$N_heads/N_flips$N
head(N_flips)## N N_heads prop
## 1 1 1 1.0000000
## 2 2 2 1.0000000
## 3 3 1 0.3333333
## 4 4 3 0.7500000
## 5 5 1 0.2000000
## 6 6 1 0.1666667If we look at the first 100, you can see
ggplot(data = N_flips) +
geom_line(aes(x = N, y = prop)) +
geom_hline(aes(yintercept = 0.5), colour = "red", linetype = "dotted") +
coord_cartesian(xlim = c(0, 100)) +
theme_bw()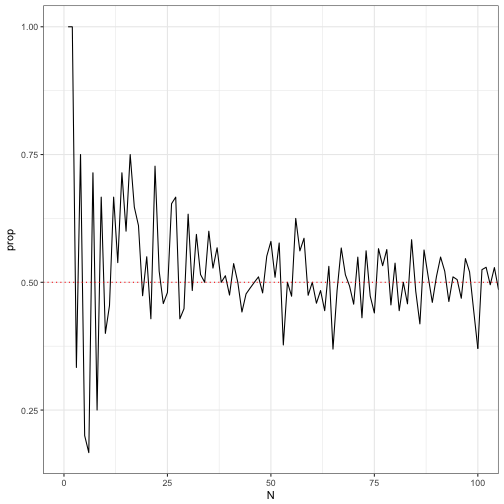
Lets look at the whole range
ggplot(data = N_flips) +
geom_line(aes(x = N, y = prop)) +
geom_hline(aes(yintercept = 0.5), colour = "red", linetype = "dotted") +
coord_cartesian(xlim = c(0, 5000)) +
theme_bw()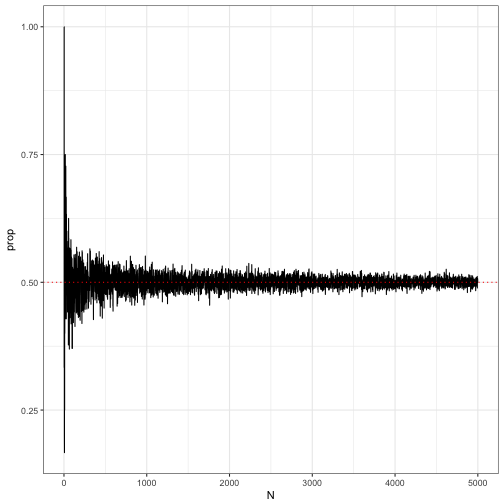
-
The Bayesian interpretation of probability is the degrees of belief. For a Bayesian, \( Pr{A}\) is a measure of certainty; a quantication of an investigator’s belief that A is true.
-
Differences in Frequentist and Bayesian perspectives are most important pertaining to inferential procedures (e.g., parameter estimation and hypothesis testing). They are irrelevant to the mathematical principles of probability.